Abstract
Background: Diffuse large B cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma (NHL), and overall, 60% of patients have achieved complete remission (CR) with standard chemotherapy with rituximab. However, we did not find the exact answers to which molecular mechanisms influence the DLBCL development and why over 40% of patients did not achieve a CR. Recently, it has been reported that the gut microbiome is deeply related to immune system modulation, as well as, these associations can influence cancer development or the quality of chemotherapy efficacy. Thus, we performed a stool analysis to assess whether differences in gut microbial composition between healthy groups and DLBCL patients are likely to influence the DLBCL development. Furthermore, we discussed whether microbial dysbiosis or specific microbes might affect treatment efficacy and safety.
Methods: Since March 2019, we have collected microbial information from 93 feces samples taken from the treatment naïve DLBCL patients. When we compared gut microbes between the healthy group and the DLBCL patients, we used the gut microbiome data from the healthy group, which had already been analyzed. All fecal samples were collected with a stool sampling kit (Chunlab Inc, Seoul, Korea) for microbial community analysis. The species richness was assessed using the Chao1, and evenness was calculated using the Shannon index. The beta-diversity was calculated with the Bray-Curtis metric and visualized with principal coordinate analysis (PCoA). The linear discriminant analysis effect size (LEfSe) was employed to determine the taxa and inferred pathways associated with one of the two groups to identify the potential biomarkers. The variation in taxonomic profiles and diversity indices between patients and healthy controls were analyzed using the Mann-Whitney U test and permutational multivariate analysis of variance (PERMANOVA). We used a stringent logarithmic, linear discriminant analysis (LDA) score threshold of 3.0 for taxonomic and functional markers for the LEfse outputs.
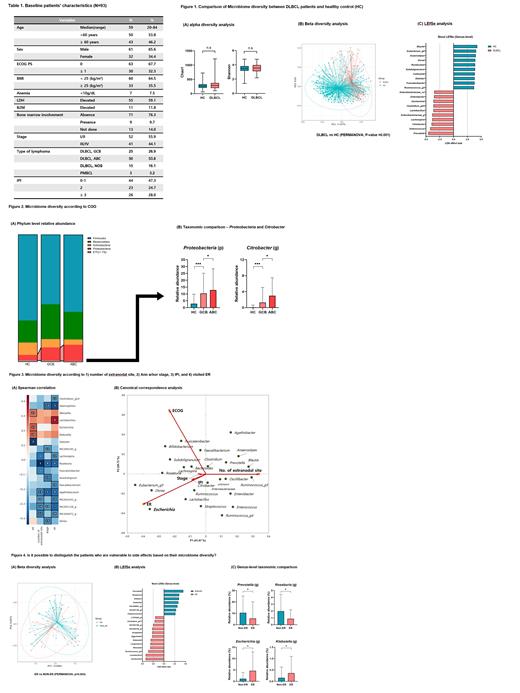
Results: Among the 93 patients, 41 patients (44.1%) had stage III or IV disease. According to the alpha diversity analysis, there were no differences in richness (Chao1) and evenness (Shannon) between DLBCL patients and the healthy control group (Fig 1A). However, we recognized that the microbial composition between these two groups was significantly different (PERMANOVA, P-value=0.001, Fig 1B). We found increased pathogenic Enterobacteriaceae genera, such as Enterococcus, Citrobacter, Escherichia, Enterobacter, and Sutterella, were markers of gut dysbiosis in DLBCL patients (Fig 1C). Furthermore, ABC subtype of DLBCL showed higher proteobacteria at the phylum level and Citrobacter at the genus level than GCB subtype (Fig 2A,B). Besides, genus Escherichia, Klebsiella, Lactobacillus, and Weissella showed a positive correlation with the indicators predicted to be related to the disease burden (number of extranodal sites, Ann Arbor stage, international prognostic index) and the side effects (admission or visit in the emergency room [ER] due to any chemotherapy-related toxicities) estimated by Spearman correlation and canonical correspondence analysis (Fig 3A, B). Additionally, there was a significant difference in intestinal microbes composition between patients who experienced the side effects or not (PERMANOVA, P-value=0.003, Fig 4A). Furthermore, facultative anaerobe and opportunistic pathogens, such as genus Escherichia, Klebsiella, Lactobacillus, and Weissella, were abundant in patients who experienced the side effects. These results suggested that patients with severe dysbiosis are more susceptible to the side effects of chemotherapy (Fig 4B, C).
Conclusion: Through these results, it was possible to know the specific intestinal microbiome distribution of DLBCL. Also, we found an abundance of opportunistic pathogens (Escherichia, Klebsiella, Lactobacillus, and Weissella) in DLBCL patients with a high disease burden or some susceptible features about chemotherapy. However, we presented the intestinal microbes configuration and did not prove the mechanism by which the gut microbes affect the tumor environment related to lymphoma evolution. Therefore, we expect that the currently planned functional study of specific DLBCL-related microbes and diversity will answer this question.
Kim: Roche: Research Funding; Sanofi: Research Funding; IGM Biosciences: Research Funding; Celltrion: Research Funding; Dong-A Pharmaceutical: Research Funding; Eisai: Research Funding; Johnson & Johnson: Research Funding; Kyowa Kirin: Research Funding.